In this NewsFlash, we find out why our model of the circadian clock might be wrong, how the bacteria that live inside a cow's gut could help make better biofuels, and why groups of animals make better decisions. Plus, how some bacteria make rapid changes to their genome - avoiding the immune system as well as preventing us from making effective vaccines.
In this episode
00:27 - Resetting the Circadian Clock
Resetting the Circadian Clock
Biological clocks play an essential role in physiology and in controlling behaviour - from regulating sleep cycles in animals to balancing photosynthesis in plants. Now, research published in Nature suggests that our model of how the clock works might be wrong.
There are two key features that define a biological clock mechanism: It must compensate for temperature, as chemical reactions will generally run faster at higher temperatures; and it must be able to synchronise to external stimuli such as light and dark.
 Current models of circadian clocks rely heavily on feedback loops based around transcription and translation - the "reading" of DNA and subsequent protein production. Essentially, the products of transcribing "clock" genes regulate how associated genes are expressed, and this leads to a roughly 24 hour feedback loop. Post-transcription contributions to the circadian clock have been observed, but the basic DNA-led feedback mechanism is still assumed to be the major driving force in biological clocks.
Current models of circadian clocks rely heavily on feedback loops based around transcription and translation - the "reading" of DNA and subsequent protein production. Essentially, the products of transcribing "clock" genes regulate how associated genes are expressed, and this leads to a roughly 24 hour feedback loop. Post-transcription contributions to the circadian clock have been observed, but the basic DNA-led feedback mechanism is still assumed to be the major driving force in biological clocks.
Now, John O'Neill & Akhilesh Reddy at Addenbrookes Hospital in Cambridge have shown that non-transcriptional mechanisms are capable of maintaining a clock in human red blood cells, which have no nucleus and are therefore incapable of transcription.
To study clocks in red blood cells, they looked to a family of antioxidant proteins called Peroxiredoxins or PRX. These are responsible for clearing reactive oxygen species like peroxide from the cell. A subclass of PRX molecules undergo an oxidation/reduction, also known as a "redox" reaction, with a regular cycle.
To assess PRX's suitability as a clock molecule, O'Neill and Reddy took red blood cell samples from healthy volunteers, and stored them in darkness and at a constant temperature. This enabled them to sample the cells every 4 hours, and confirm that the redox cycle did indeed take around 24 hours.
To see if this system fulfilled the other conditions of being a clock molecule, they attempted to entrain the cells to a cycle of high and low temperature - mirroring the temperature variations you or I might experience on a daily basis. After just 48 hours, the PRX redox cycle was seen to sync with the new temperature regime, making it an excellent candidate for non-transcription based circadian rhythm control.
PRX proteins are very highly conserved, and found in a huge range of species, including mammals, plants and algae. In a related paper, Andrew Millar from the University of Edinburgh, along with colleagues in Cambridge and France, show the same mechanism in action in Ostreococcus tauri, a single celled, eukaryotic alga. This raises some very interesting questions about our current understanding of circadian clocks, and raises some exciting prospects for our understanding of clock evolution.

03:20 - Chewing the cud (and how it got there)
Chewing the cud (and how it got there)
Researchers in America this week have dramatically enlarged the catalogue of known genes which allow an organism to break down plants. This new information could be very useful in producing biofuel from plant matter.
 Matthias Hess and colleagues found their new biomass-breaking genes and genomes swimming around in the rumen of a cow. The rumen is a fascinating part of the cow's digestive system in which lots of bacteria, yeasts and fungi live. These microbes break down all the grass and leaves that the cow munches through - so as you can imagine, a probiotic yoghurt means nothing to the host animals.
Matthias Hess and colleagues found their new biomass-breaking genes and genomes swimming around in the rumen of a cow. The rumen is a fascinating part of the cow's digestive system in which lots of bacteria, yeasts and fungi live. These microbes break down all the grass and leaves that the cow munches through - so as you can imagine, a probiotic yoghurt means nothing to the host animals.
Publishing in Science, Hess and his team were able to sequence a quarter of a terabase (which is an enormous amount - a trillion bases) of genomic information from many different microbes involved in digestion. They were also able to identify the very genes which were involved in plant degradation and the proteins which are thought to do the work.
Just in terms of the data generated, it's quite a step forward in sequencing large communities of different microbes. And the team have managed to demonstrate that it's possible to construct genomes of new, previously unknown organisms from the same mass of data. But the authors hope that this work will ultimately help other researchers in developing more efficient ways of producing biofuels.
04:42 - Rapid Change in Bacterial Genome
Rapid Change in Bacterial Genome
Dr Stephen Bentley, Wellcome Trust Sanger Institute
Ben - Also this week, researchers at the Wellcome Trust Sanger Institute who have been looking at changes in the genome of a troublesome pathogen, Streptococcus pneumoniae. This bacteria is responsible for a broad range of human diseases including pneumonia, ear infections, and bacterial meningitis. Fortunately, because this bacteria has been infecting humans for so long, we have samples from all over the world, going back many, many years, and this means, we can compare them genetically to see what has changed in response to modern antibiotic's and to vaccines. We're joined now by Dr. Stephen Bentley from the Wellcome Trust Sanger Institute. Thank you very much for joining us, Stephen.
Stephen - My pleasure.
 Ben - Now the idea that bacteria change genetically in response to drugs or vaccines isn't new, we've known about this for a while. What's novel about your work?
Ben - Now the idea that bacteria change genetically in response to drugs or vaccines isn't new, we've known about this for a while. What's novel about your work?
Stephen - As you say, we've known about these phenomena occurring over time since antibiotics have been in use, but the kind of breakthrough that we have in the projects that we're now able to do has been driven by our ability to sequence large number of genomes. Ten years ago, when we tried, when we sequenced the genomes of things like TB, Mycobacterium tuberculosis, one project to sequence the genome of one isolate, which was chosen to represent species, would have taken a couple of years and cost 1 or 2 million Pounds. The new sequencing technologies allow us to sequence hundreds or thousands of isolates at a cost of around 100 Pounds per isolate, and the turnaround of generating sequence data is down to a matter of weeks. That allows us now to exploit the collections of isolates that you mentioned earlier to really drill down onto the evolution of a population.
Ben - So, what samples have you actually been looking at? I understand they are from all over the world and actually going back quite a long time.
Stephen - The idea of project was to really use the whole genomes to analyse the evolution over the period since the introduction of antibiotics. So, when we tried to collect the samples we were looking for as much geographic and temporal coverage as possible. So, the collection we ended up with was 240 isolates, spanning the period of 1984 to 2007, and covering 22 different countries around the globe.
Ben - What sort of scale of changes are you actually seeing in the genome here?
Stephen - Substitution mutations which basically occur at random. One of those will occur approximately every 15 weeks. But there's another mechanism for variation which we call recombination, and that's basically where the bacteria are able to swap DNA with their neighbours. On the BBC website, there's a guy, James Gallagher, who's come up with a really nice analogy that's like doing down to the shops and swapping eye colour with someone in the queue at the checkout. But for the bacteria, that means they can generate enormous amounts of variation in their genomes and each variation that's generated then can be selected for possible advantage. So in a situation where the organisms are being exposed to antibiotics on a regular basis, which is what's happened in all bacteria since the introduction of antibiotics in the '60s and '70s, if they acquire a variation in their sequence which gives them advantage over the rest, then that's going to give them a good chance of proliferating.
Ben - Now obviously, the bacteria can't, as you say, meet down the shops and have a chat and say, "I've got these bright blue eyes. They've been really useful for me for survival. Why don't you have a copy?" There can't be a process by which they know that if they translate these genes from another cell that'll be useful. How does that process actually work? Is it quite random?
Stephen - It is entirely random. So, what happens with Streptococcus pneumoniae, they live in the human nasopharynx and, as far as we know, that's their only natural niche, but there are other bacteria that live there, some of which will be members of the same species but maybe only slightly-related, and then there are other species as well. As cells grow and divide and go through a cycle where there's actually death, some cells will lyse. Lysing the cells releases the [DNA] bacteria into the environment and that allows bacteria like Strep pneumo to potentially take-up that DNA. So this ability to take-up DNA from the environment is not ubiquitous in bacteria, it only happens in certain species. But Strep pneumo does that and is entirely random and some people have thought about whether it might actually be for nutritional needs and much as anything that they take up the DNA. So, the randomness is there but then selection kicks in, so it's really - if the variation you introduce is disadvantageous, then that will very quickly die out. If it happens to give you an advantage, then it will proliferate.
Ben - With all these random changes, do you see changes evenly across the genome or are there regions that are very heavily conserved and regions that are very highly variable?
Stephen - Yes. So we see these variation hotspots in the genome and it's interesting that those variation hotspots tend to be associated with surface antigens. So the major surface antigen in Strep pneumo is the surface polysaccharide and that's one region where we've seen frequent recombinations.
Ben - Now these are the sugars or the proteins that are on the surface of the cell that it actually presents to, say, our immune system.
Stephen - Exactly, yes. And what's interesting from the data that we've seen is that that region is a hotspot, so it's swapping in and out with its neighbours and changing its surface polysaccharide. But these changes were already in the population at fairly high levels, so that in around 2000, when we started to introduce a vaccine which targeted those surface polysaccharides, we could see that, in the population, there were already variants generated which we're going to be able to evade that vaccine because they didn't have the vaccine target.
Ben - So they essentially come preloaded to avoid our vaccine attempts.
Stephen - The population does, yes.
Ben - How can this help us to develop better vaccines or better antibiotics?
Stephen - Already, since the seven-valent vaccine was introduced, because these capsules have been spotted, new generations of vaccine have been generated where they now target 10 or 13 of those types. So, going forward, we would hope to be able to continue to monitor the population in very, very high resolution, using whole genome sequencing, so that we'll be able to understand better how the pathogens are responding to the clinical practices; and then maybe we can adjust those clinical practices to make them more efficient.

11:43 - Many heads are better than one
Many heads are better than one
Animals come to better decisions more quickly in larger groups, according to research published in the journal PNAS this week.
Group decision making is seen in many social communities - from ants to humans. It forms the basis of financial markets and the behaviour of the internet. It's clearly evolved many times over, but exactly how the decision making process in a group differs from that an individual is hard to determine.
 An international team of researchers, including Ashley Ward at the University of Sydney and Jens Krause at Humboldt University in Berlin, created a simple decision making task - a Y shaped chamber where one path contains a replica predator, and the other was safe. They then introduced mosquitofish, Gambusia holbrooki, to the task either alone, in pairs or groups of 4, 8 or 16.
An international team of researchers, including Ashley Ward at the University of Sydney and Jens Krause at Humboldt University in Berlin, created a simple decision making task - a Y shaped chamber where one path contains a replica predator, and the other was safe. They then introduced mosquitofish, Gambusia holbrooki, to the task either alone, in pairs or groups of 4, 8 or 16.
They then monitored how the fish moved, and which direction they chose to take, to see if group size would alter the speed and accuracy of the decision making process.
They found that lone fish took longer to reach a decision, swimming more slowly and changing direction often, but even then only chose the "safe" route little over half the time. As the group size increased, the decisions became more accurate and much faster - the larger group choosing the safe route up to 90% of the time, despite swimming faster and taking a more direct route.
The next step was to try to work out why groups are so much better at these decisions. As the variation between different lone fish was insignificant, this rules out the idea that particular fish are simply "experts" at spotting predators, benefiting the rest of the group. There is some evidence that the group does divides the responsibility of being vigilant - each fish needs to scan a smaller area, so the amount of direction changes made reduces in larger groups. Task sharing like this also means the fish must rely on social cues to make these decisions, as was seen when observing the fish closest to the leading fish.
This research suggests a high degree of cooperation, division of labour and rapid communication between members of a group, all factors that would promote the evolution of group decision making.
14:36 - Brain activity can demonstrate how fast a second language is learned
Brain activity can demonstrate how fast a second language is learned
This week teams in both Hong Kong and Chicago have found that the levels of activity in two specific areas of the brain can be used to predict how well someone is learning a second language.
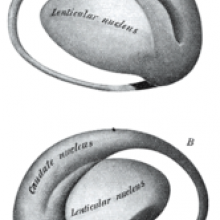
 Li Hai Tan and colleagues found that the left caudate and fusiform gyrus areas of the brain displayed more activity the better the language student performed in their tests. The fusiform part of the brain is located at the back and base of the brain while the caudate is found closer to the centre and is shaped a bit like hooked headphones.
Li Hai Tan and colleagues found that the left caudate and fusiform gyrus areas of the brain displayed more activity the better the language student performed in their tests. The fusiform part of the brain is located at the back and base of the brain while the caudate is found closer to the centre and is shaped a bit like hooked headphones.
They used fMRI (functional magnetic resonance imaging) in between tests taken by 26 10 year old Chinese students who were learning English. They were given the first written test just before the fMRI scan and the second one year later. During the fMRI scan the children were asked to identify written English words. They found that those who'd demonstrated the greatest amount of activity in the caudate and fusiform regions during the scan performed better in the first and second tests.
Publishing in the journal PNAS, Tan and his team believe that the amount of activity in these two brain areas can therefore be used to predict how well a student will do when they learn a second language.










Comments
Add a comment